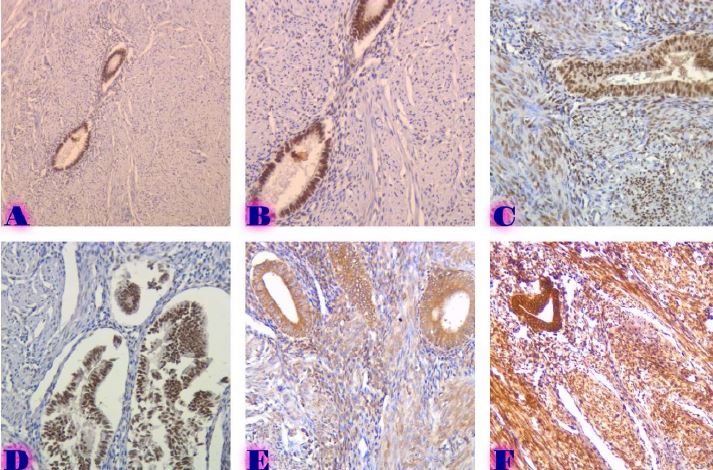
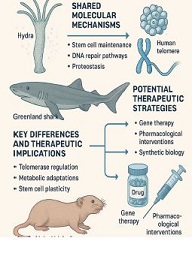
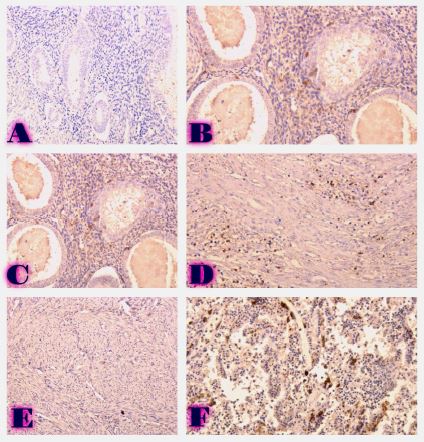
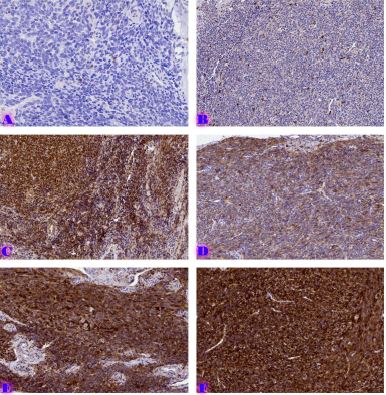
BIOINFORMATICS APPROACHES FOR MULTI-OMICS ANALYSIS OF THE TUMOR MICROENVIRONMENT: INTEGRATING PROTEOMICS, EXOSOMES AND IMMUNE SIGNALING

Downloads
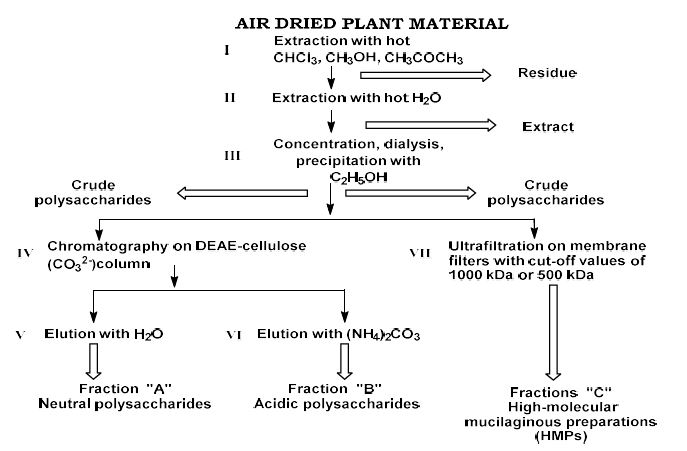
Cancer stem cell (CSC) plasticity is increasingly recognized as a central driver of tumor initiation, therapeutic resistance, metastatic dissemination, and disease recurrence. Yet, the dynamic transitions between stem-like and differentiated malignant states remain poorly resolved due to the intrinsic heterogeneity of tumors and the limitations of bulk profiling methods. Recent advances in single-cell sequencing technologies have created an unprecedented opportunity to dissect CSC states at high resolution; however, extracting mechanistic insights from these data requires sophisticated computational methods.
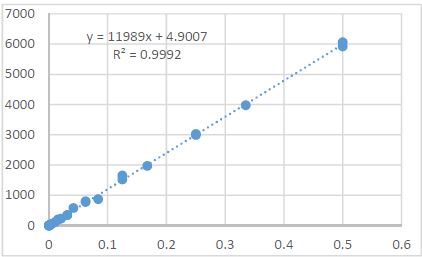
In this work, we present an integrative bioinformatic framework for characterizing CSC plasticity across diverse cancer types using single-cell transcriptomic, epigenomic, and proteogenomic datasets. Our approach synthesizes multiple layers of information—including transcriptional variability, regulatory network inference, cell trajectory reconstruction, chromatin accessibility, ligand–receptor communication, and lineage mapping—to identify key determinants that regulate transitions between stem-like, progenitor-like, and differentiated malignant phenotypes. Through integrative clustering, pseudotime analysis, and machine-learning–based state prediction, we uncover conserved molecular programs and dynamic regulatory modules that define CSC behavior.
Furthermore, we implement cell–cell communication modeling to map how CSC niche interactions influence plasticity, focusing on immune–CSC crosstalk, stromal signaling circuits, and exosome-mediated molecular transport. The combined analysis highlights novel candidate biomarkers and therapeutic vulnerabilities associated with reversible stemness states.
Together, this study demonstrates that integrated single-cell bioinformatic analysis provides a powerful framework to decode cancer stem cell plasticity, enabling a deeper mechanistic understanding of tumor evolution and identifying new avenues for precision oncology and targeted therapeutic interventions.
Downloads
Aibar, S., González-Blas, C. B., Moerman, T. (2017). SCENIC: single-cell regulatory network inference and clustering. Nature Methods, 14(11), 1083–1086.
Argelaguet, R., Arnol, D., Bredikhin, D., Deloro, Y., Velten, B., Marioni, J. C., & Stegle, O. (2020). MOFA+: a statistical framework for comprehensive integration of multi-modal single-cell data. Genome Biology, 21(1), 111.
Balázsi, G., van Oudenaarden, A., & Collins, J. J. (2011). Cellular decision making and biological noise: from microbes to mammals. Cell, 144(6), 910–925.
Batlle, E., & Clevers, H. (2017). Cancer stem cells revisited. Nature Medicine, 23(10), 1124–1134.
Batlle, E., Sancho, E., Francí, C., Domínguez, D., Monfar, M., Baulida, J., & García De Herreros, A. (2000). The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nature Cell Biology, 2(2), 84–89.
Becker, A., Thakur, B. K., Weiss, J. M., Kim, H. S., Peinado, H., & Lyden, D. (2016). Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell, 30(6), 836–848.
Benci, J. L., Xu, B., Qiu, Y., Wu, T. J. (2016). Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell, 167(6), 1540–1554.e12.
Bergen, V., Lange, M., Peidli, S., Wolf, F. A., & Theis, F. J. (2020). Generalizing RNA velocity to transient cell states through dynamical modeling. Nature Biotechnology, 38(12), 1408–1414.
Boumahdi, S., & de Sauvage, F. J. (2020). The great escape: tumour cell plasticity in resistance to targeted therapy. Nature Reviews Drug Discovery, 19(1), 39–56.
Browaeys, R., Saelens, W., & Saeys, Y. (2020). NicheNet: modeling intercellular communication by linking ligands to target genes. Nature Methods, 17(2), 159–162.
Buenrostro, J. D., Wu, B., Chang, H. Y., & Greenleaf, W. J. (2015). ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Current Protocols in Molecular Biology, 109(1), 21.29.1–21.29.9.
Carette, D., Gillard, M., Tost, J., & Frommer, M. (2019). TGF-β and cancer-associated fibroblasts in the breast tumor microenvironment. Journal of Mammary Gland Biology and Neoplasia, 24, 63–75.
Cassetta, L., & Pollard, J. W. (2018). Targeting macrophages: therapeutic approaches in cancer. Nature Reviews Drug Discovery, 17(12), 887–904.
Cassetta, L., & Pollard, J. W. (2020). Tumor-associated macrophages. Current Biology, 30(6), R246–R248.
Chaffer, C. L., Marjanovic, N. D., Lee, T., Bell, G., Kleer, C. (2013). Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell, 154(1), 61–74.
Chevrier, S., Levine, J. H., Zanotelli, V. R. T., Silina, K., Schulz, D., Bacac, M., Ries, C. H., Ailles, L., Jewett, M. A. S., Moch, H., van den Broek, M., Beisel, C., Stadler, M. B., Gedye, C., Reis, B., Pe'er, D., & Bodenmiller, B. (2018). An immune atlas of clear cell renal cell carcinoma. Cell, 169(4), 736–749.e18.
Corces, M. R., Granja, J. M., Shams, S. (2018). The chromatin accessibility landscape of primary human cancers. Science, 362(6413), eaav1898.
Cusanovich, D. A., Hill, A. J., Aghamirzaie, D., Daza, R. M.(2018). A single-cell atlas of in vivo mammalian chromatin accessibility. Cell, 174(5), 1309–1324.e18.
Dongre, A., & Weinberg, R. A. (2019). New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nature Reviews Molecular Cell Biology, 20(2), 69–84.
Efremova, M., Vento-Tormo, M., Teichmann, S. A., & Vento-Tormo, R. (2020). CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nature Protocols, 15(4), 1484–1506.
Falletta, P., Sanchez-del-Campo, L., Chauhan, J., Effern, M., Kenyon, A., Kershaw, C. J., Siddaway, R., Lisle, R., Freter, R., Daniels. (2017). Translation reprogramming is an evolutionarily conserved driver of phenotypic plasticity and therapeutic resistance in melanoma. Genes & Development, 31(1), 18–33.
Faubert, B., Solmonson, A., & DeBerardinis, R. J. (2020). Metabolic reprogramming and cancer progression. Science, 368(6487), eaaw5473.
Finnerty, B., Kleemann, S., Lei, X., King, M. R., Zhao, T., Hernandez, J. M., & Zarnegar, R. (2022). Somatic chromatin remodeling mutations drive malignant transformation of thyroid nodules. Endocrine-Related Cancer, 29(5), 269–278.
Francis, J. M., Zhang, C. Z., Maire, C. L., Jung. (2014). EGFR variant heterogeneity in glioblastoma resolved through single-nucleus sequencing. Cancer Discovery, 4(8), 956–971.
Glodde, N., Bald, T., van den Boorn-Konijnenberg, D., Nakamura, K., O'Donnell, J. (2017). Reactive neutrophil responses dependent on the receptor tyrosine kinase c-MET limit cancer immunotherapy. Immunity, 47(4), 789–802.e9.
Grivennikov, S. I., Greten, F. R., & Karin, M. (2010). Immunity, inflammation, and cancer. Cell, 140(6), 883–899.
Hangauer, M. J., Viswanathan, V. S., Ryan, M. J.(2017). Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature, 551(7679), 247–250.
Hao, Y., Hao, S., Andersen-Nissen, E. (2021). Integrated analysis of multimodal single-cell data. Cell, 184(13), 3573–3587.e29.
Herman, J. S., Sagar, & Grün, D. (2018). FateID infers cell fate bias in multipotent progenitors from single-cell RNA-seq data. Nature Methods, 15(5), 379–386.
Iacono, G., Massoni-Badosa, R., & Heyn, H. (2019). Single-cell transcriptomics unveils gene regulatory network plasticity. Genome Biology, 20(1), 110.
Jensen, C., Madsen, D. H., Hansen, M.(2021). Non-invasive biomarkers derived from the extracellular matrix associate with response to immune checkpoint blockade (anti-CTLA-4) in metastatic melanoma patients. Journal for ImmunoTherapy of Cancer, 6(1), 152.
Jerby-Arnon, L., Shah, P., Cuoco, M. S., Rodman, C. (2018). A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell, 175(4), 984–997.e24.
Kalluri, R. (2016). The biology and function of fibroblasts in cancer. Nature Reviews Cancer, 16(9), 582–598.
Kim, N., Kim, H. K., Lee, K., Hong, Y. (2020). Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nature Communications, 11(1), 2285.
Kumar, M. P., Du, J., Lagoudas, G., Jiao, Y., Sawyer, A., Drummond, D. C., Lauffenburger, D. A., & Raue, A. (2018). Analysis of single-cell RNA-seq identifies cell-cell communication associated with tumor characteristics. Cell Reports, 25(6), 1458–1468.e4.
La Manno, G., Soldatov, R., Zeisel, A.(2018). RNA velocity of single cells. Nature, 560(7719), 494–498.
Lamouille, S., Xu, J., & Derynck, R. (2014). Molecular mechanisms of epithelial–mesenchymal transition. Nature Reviews Molecular Cell Biology, 15(3), 178–196.
Lee, H. O., Hong, Y., Etlioglu, H. E. (2020). Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nature Genetics, 52(6), 594–603.
Liau, B. B., Sievers, C., Donohue, L. K. (2017). Adaptive chromatin remodeling drives glioblastoma stem cell plasticity and drug tolerance. Cell Stem Cell, 20(2), 233–246.e7.
Lotfollahi, M., Wolf, F. A., & Theis, F. J. (2019). scGen predicts single-cell perturbation responses. Nature Methods, 16(8), 715–721.
Ma, S., Zhang, B., LaFave, L. M., Earl, A. S.(2020). Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell, 183(4), 1103–1116.e20.
Marjanovic, N. D., Hofree, M., Chan, J. E. (2020). Emergence of a high-plasticity cell state during lung cancer evolution. Cancer Cell, 38(2), 229–246.e13.
Meacham, C. E., & Morrison, S. J. (2013). Tumour heterogeneity and cancer cell plasticity. Nature, 501(7467), 328–337.
Neftel, C., Laffy, J., Filbin, M. G., Hara. (2019). An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell, 178(4), 835–849.e21.
Nguyen, P. H. D., Ma, S., Phua, C. Z. J., Kaya. (2020). Intratumoural immune heterogeneity as a hallmark of tumour evolution and progression in hepatocellular carcinoma. Nature Communications, 11(1), 227.
O'Donnell, J. S., Long, G. V., Scolyer, R. A., Teng, M. W., & Smyth, M. J. (2017). Resistance to PD1/PDL1 checkpoint inhibition. Cancer Treatment Reviews, 52, 71–81.
Ogden, A., Garlapati, C., Li, X. B., Turaga, R. C.(2020). Multi-institutional study of nuclear KIFC1 as a biomarker of poor prognosis in African American women with triple-negative breast cancer. Scientific Reports, 10(1), 7944.
Oren, Y., Tsabar, M., Cuoco, M. S., Amir-Zilberstein. (2021). Cycling cancer persister cells arise from lineages with distinct programs. Nature, 596(7873), 576–582.
Pastushenko, I., Brisebarre, A., Sifrim, A., (2018). Identification of the tumour transition states occurring during EMT. Nature, 556(7702), 463–468.
Pérez-García, A., Pérez-Jannotti, G., Sánchez-Céspedes, R. (2022). Epigenetic mechanisms in breast cancer therapy and resistance. International Journal of Molecular Sciences, 23(7), 3721.
Rambow, F., Rogiers, A., Marin-Bejar, O., Aibar, S. (2018). Toward minimal residual disease-directed therapy in melanoma. Cell, 174(4), 843–855.e19.
Reya, T., & Clevers, H. (2005). Wnt signalling in stem cells and cancer. Nature, 434(7035), 843–850.
Rheinbay, E., Nielsen, M. M., Abascal, F., Wala, J. (2020). Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature, 578(7793), 102–111.
Rodriques, S. G., Stickels, R. R., Goeva, A., Martin, C. A., Murray, E., Vanderburg, C. R., Welch, J., Chen, L. M., Chen, F., & Macosko, E. Z. (2019). Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science, 363(6434), 1463–1467.
Roth, A., Khattra, J., Yap, D., Wan, A., Laks, E.(2014). PyClone: statistical inference of clonal population structure in cancer. Nature Methods, 11(4), 396–398.
Saelens, W., Cannoodt, R., Todorov, H., & Saeys, Y. (2019). A comparison of single-cell trajectory inference methods. Nature Biotechnology, 37(5), 547–554.
Sharma, S. V., Lee, D. Y., Li, B., Quinlan, M. P. (2010). A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell, 141(1), 69–80.
Shibue, T., & Weinberg, R. A. (2017). EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nature Reviews Clinical Oncology, 14(10), 611–629.
Stoeckius, M., Hafemeister, C., Stephenson, W., Houck-Loomis, B., Chattopadhyay, P. K., Swerdlow, H., Satija, R., & Smibert, P. (2017). Simultaneous epitope and transcriptome measurement in single cells. Nature Methods, 14(9), 865–868.
Street, K., Risso, D., Fletcher, R. B., Das, (2018). Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics, 19(1), 477.
Stuart, T., Butler, A., Hoffman, P., Hafemeister, C., Papalexi, E., Mauck III, W. M., Hao, Y., Stoeckius, M., Smibert, P., & Satija, R. (2019). Comprehensive integration of single-cell data. Cell, 177(7), 1888–1902.e21.
Su, Y., Wei, W., Robert, L., Xue, M. (2017). Single-cell analysis resolves the cell state transition and signaling dynamics associated with melanoma drug-induced resistance. Proceedings of the National Academy of Sciences, 114(52), 13679–13684.
Teschendorff, A. E., & Enver, T. (2017). Single-cell entropy for accurate estimation of differentiation potency from a cell's transcriptome. Nature Communications, 8(1), 15599.
Tirosh, I., Izar, B., Prakadan, S. M., Wadsworth II, M. H.(2016). Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science, 352(6282), 189–196.
Trapnell, C., Cacchiarelli, D., Grimsby, J.(2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature Biotechnology, 32(4), 381–386.
Tsoi, J., Robert, L., Paraiso, K., Galvan, C.(2018). Multi-stage differentiation defines melanoma subtypes with differential vulnerability to drug-induced iron-dependent oxidative stress. Cancer Cell, 33(5), 890–904.e5.
Vaughan, A. E., & Chapman, H. A. (2013). Regenerative activity of the lung after epithelial injury. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease, 1832(7), 922–930.
Wolf, F. A., Angerer, P., & Theis, F. J. (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biology, 19(1), 15.
Wolf, F. A., Hamey, F. K., Plass, M., Solana, J., Dahlin, J. S. (2019). PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biology, 20(1), 59.
Wu, S. Z., Al-Eryani, G., Roden, D. L., Junankar, S., Harvey. (2021). A single-cell and spatially resolved atlas of human breast cancers. Nature Genetics, 53(9), 1334–1347.
Zafar, H., Wang, Y., Nakhleh, L., Navin, N., & Chen, K. (2019). Monovar: single-nucleotide variant detection in single cells. Nature Methods, 16(7), 585–587.
Zhang, M., Eichhorn, S. W., Zingg, B., Yao, Z., Cotter, K.(2021). Spatially resolved cell atlas of the mouse primary motor cortex by MERFISH. Nature, 598(7879), 137–143.
Zheng, G. X. Y., Terry, J. M. (2017). Massively parallel digital transcriptional profiling of single cells. Nature Communications, 8(1), 14049.
Aphkhazava, D., Tuphinashvili, T., Sulashvili, N.(2023). The Features and Role of SHP2 Protein in Postnatal Muscle Development. Scientific Journal „Spectri“, 1. https://doi.org/10.52340/spectri.2023.01
Sulashvili N, Davitashvili M, Gorgaslidze N, Gabunia L. The scientific discussion of some issues of features and challenges of using of car-t cells in immunotherapy. GS 2024 ;6(4):263-90. https://journals.4science.ge/index.php/GS/article/view/3214
Aphkhazava, David, Nodar Sulashvili, and Jaba Tkemaladze. 2025. “Stem Cell Systems and Regeneration”. Georgian Scientists 7 (1):271-319. https://doi.org/10.52340/gs.2025.07.01.26.
Relling, Mary V., and William E. Evans. 2015. “Pharmacogenomics in the Clinic.” Nature 526 (7573): 343–50.
Aphkhazava, David, Tamar Tuphinashvili, Nodar Sulashvili. 2023. “THE FEATURES AND ROLE OF SHP2 PROTEIN IN POSTNATAL MUSCLE DEVELOPMENT”. Scientific Journal „Spectri“ 1 (March). https://doi.org/10.52340/spectri.2023.01.
Aphkhazava, David, Nodar Sulashvili, Ia Egnatievi, Manana Giorgobiani. 2024. “Derivation of AMPA Receptor GluA1 Subunits in Mice from Exosomes Modulates Inflammatory Pain”. Scientific Journal „Spectri“ 9 (1). https://doi.org/10.52340/spectri.2024.09.01.05.
Aphkhazava, David, Nodar Sulashvili, Ia Egnatievi, Tamar Tupinashvili. 2024. “DYNAMIC TUMOR MICROENVIRONMENT THEORY: A MULTIFACETED APPROACH TO TUMOR RESEARCH AND BIOCHEMISTRY”. Scientific Journal „Spectri“ 9 (1). https://doi.org/10.52340/spectri.2024.09.01.06.
Qiu, X., Zhang, Y., Martin-Rufino, J. D., (2022). Mapping transcriptomic vector fields of single cells. Cell, 185(4), 690–711.e45.
Quintanal-Villalonga, Á., Chan, J. M., Yu, H. (2020). Lineage plasticity in cancer: a shared pathway of therapeutic resistance. Nature Reviews Clinical Oncology, 17(6), 360–371.
Raj, A., Rifkin, S. A., Andersen, E., & van Oudenaarden, A. (2021). Variability in gene expression underlies incomplete penetrance. Nature, 463(7283), 913–918.
Copyright (c) 2025 Georgian Scientists

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.